State-of-the-art advanced imaging platform, custom-built microscopes, and advanced 3D histology

Leading the future of neuroscience discovery
Combining the expertise of a multidisciplinary team, including scientists, engineers, and neurobiologists, we are developing new technologies, cell-targeted gene therapies, coupled with implantable, bio-compatible optoelectronic devices.
Our expertise includes:
- Molecular biology, histology, genetic engineering, cell culture (BSL1 and BSL2) contributing to our expansion into the field of neurobiology.
- Adanced microscopy-imaging.
- Brain mapping: tissue processing, 3D multiplexlabeling, hardware, computational analysis and image processing.

3D Tissue profiling and Spatial Biology
Our team is developing cutting-edge tissue profiling techniques to map the molecular and cellular landscapes of the brain. By characterizing both healthy and diseased states, we aim to predict disease evolution and deepen our understanding of neurological conditions.
To achieve this, we are advancing tissue processing pipelines that preserve spatial context and enhance resolution. These include:
- Whole-organ clearing and labeling to maintain tissue integrity, uncover neuronal connections, and reveal intricate vascular networks and rare cell populations.
- Multiplexed and multi-omics RNA and protein staining for comprehensive molecular profiling.
- Tissue expansion microscopy to achieve higher spatial resolution, enabling the visualization of fine subcellular structures.
We are also developing sophisticated image processing pipelines to handle large-scale datasets, ensuring efficient analysis and extraction of meaningful insights from high-resolution imaging data.
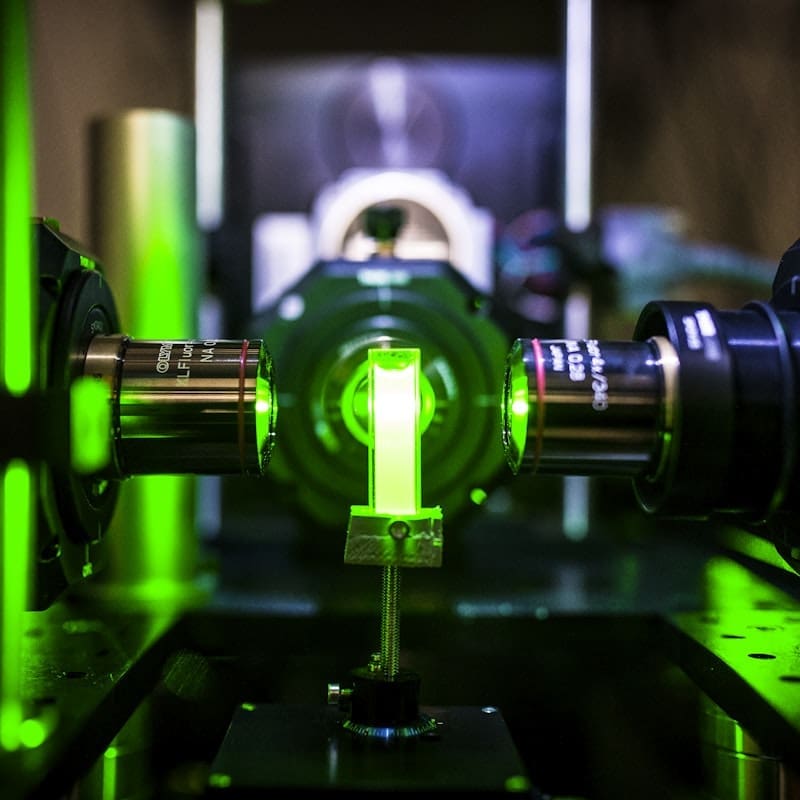
Lightsheet microscopes can reveal the 3D anatomy of entire small organs. They image brain tissue down to individual neurons and offer unprecedented maps of nervous system structure and function. ALICe, the Advanced Lightsheet Imaging Center, integrates a series of innovative fluorescence microscopy tools in a single pipeline to image whole organs with custom-built microscopes at high spatial and temporal resolution.

The multidisciplinary ALICe team unites expertise from physics, cell biology, neuroscience and engineering, and joins forces with research teams around the world to image and draw insights into the central and peripheral nervous systems, innervation of organs including the brain and heart as well as human brain samples. Another focus area is brain and spinal cord organization for researchers working to restore movement after paralysis or to investigate neuronal networks involved in cognition, pleasure and drug addiction.
Unlike traditional microscopy in which specimens are cut in slices with a blade before being viewed on a slide under a microscope, lightsheet microscopes optically slice samples with a sheet of light. This optical sectioning captures slivers of image without damaging the sample. The images are then combined to reconstruct a detailed three-dimensional image of a whole organ or specimen.
The ALICe pipeline
The theoretical basis of lightsheet microscopy was established more than 100 years ago by R. A. Zsigmondy, an Austrian-Hungarian chemist[1]. His idea was to direct light through a slit, creating a lightsheet to illuminate a sample.
In the early 2000s, Jan Huisken and collaborators demonstrated for the first time that this approach could be used to image biological samples at sub micrometer spatial resolution, with micrometer scale optical sectioning in millimeter thick tissue[2]. Since that time and thanks to major advances in the field of tissue clarification, numerous types of lightsheet microscopes have emerged.
At one end of the spectrum is the family of high spatial resolution microscopes used either to image small transparent biological samples at high speed and extremely high spatial resolution (using multiplexed Bessel beams: Lattice lightsheet microscopy[4]) or for centimeter-sized clarified tissues at submicronic in-plane resolution (CLARITY-optimized light-sheet microscopy (COLM)[3]) or with sub-micronic isotropic resolution (Axially Swept Lightsheet Microscopy ctASLM[5]).
At the other end of the spectrum, are the mesoscale lightsheet microscopes which can image centimeter-sized samples at high speed while preserving subcellular resolution (mesoscale selective plane illumination microscopy – mesoSPIM[6]).
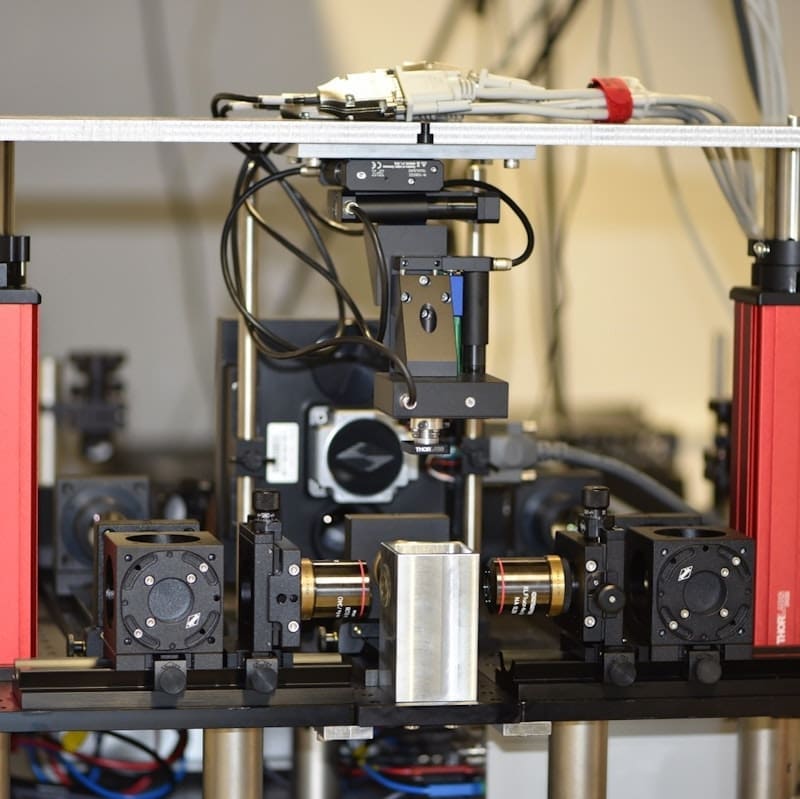
Here at the Advanced Lightsheet Imaging Center, we have implemented a high spatial resolution lightsheet microscope (COLM) which is complemented by a high speed mesoSPIM. A scientific advisory board constituted by Prof Lüscher, Prof. Courtine and Prof. Holtmaat followed and contributed for the development of ALICe’s project.
[1] H. Siedentopf and R. Zsigmondy, Ann. Phys. (Berlin), 10, 1–39 (1903).
[2] J. Huisken et al., Science, 305, 5686 (2004).
[3] R. Tomer et al., Cell, 163, 1796–1806 (2015).
[4] B-C. Chen et al., Science, 346 (2014)
[5] T. Chakraborty et al., Nat. Methods, 16 (2019)
[6] F.F. Voigt et al, Nat. Methods, 16 (2019)
Lightsheet microscope samples are made completely transparent before they are imaged. This process, known as clearing, removes all opaque lipids including fats, present in brain tissue and renders the sample transparent.
ALICe tissue clearing is predominantly based on the CLARITY protocol, with additional experience in DISCO-based and any other published clearing protocols.
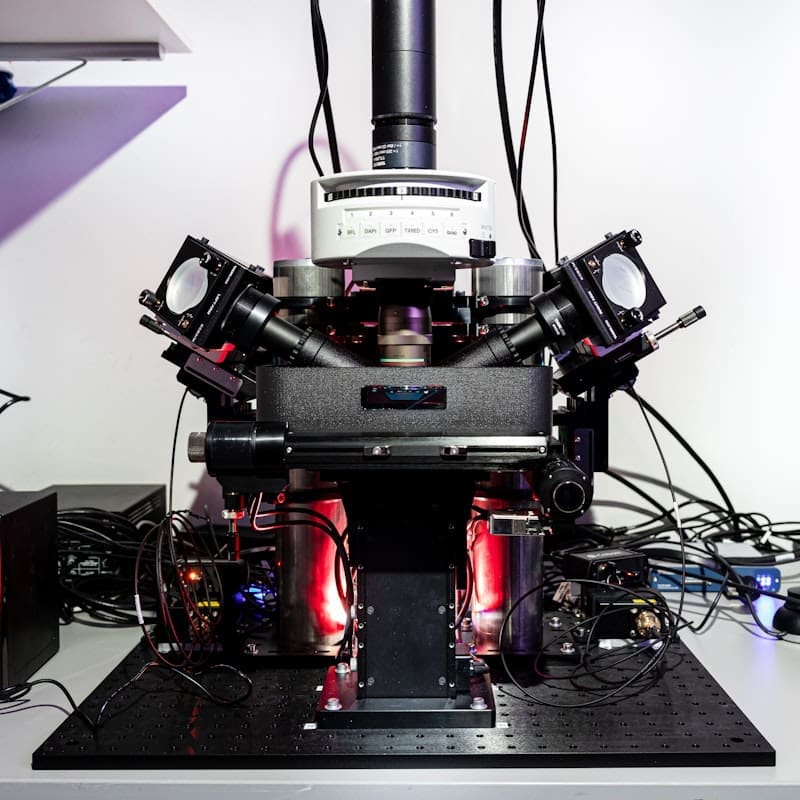
ALICE is equipped with three lightsheet systems tailored for imaging large, cleared samples. The custom-built mesosSPIM microscope is optimized for large scale screening of multiple samples at cellular resolution with high speed in imaging performances.
Clarity Optimized Light-sheet Microscope (COLM) offers the unique opportunity to capture high spatial resolution (near diffraction-limited lateral resolution), whole brain fluorescence images without perturbing neuronal architecture.
Clearscope, from MBF is light sheet theta microscope designed to meet the unique challenges inherent to imaging at high spatial resolution and reconstruct entire human cleared organs. With its theta configuration, the clearscope is able to accommodate 15×10 cm wide samples.
Finally, our confocal systems are equipped with modules for cleared tissue section imaging.
The ALICe image analysis workstations are equipped with custom-software and industry-leading software solutions including Imaris, Arivis, Matlab, ImageJ and Terastitcher to enable efficient access, extraction and analysis of 3D data from lightsheet imaged samples.
The final step of the ALICe pipeline includes 3D data visualization and analysis in a Syglass virtual reality environment to allow interaction with samples in an immersive way, speeding up the analysis process.
Seeing the invisible

Available resources
Clarity Optimized Light-sheet Microscope (COLM) – developed in collaboration with the original inventors of the technology – Raju Tomer, Columbia University and Karl Deisseroth, Stanford University – is optimized for large scale clarified tissues and allows a fast exchange of optics and samples. It offers the unique opportunity to capture high spatial resolution (near diffraction-limited lateral resolution), whole brain fluorescence images without perturbing neuronal architecture.
- Objectives: Detection: 25x/1(CLARITY) WD 8mm ; 10x/0,6 (CLARITY) WD 8 mm; : 4x/0,28 WD 29 mm; Illumination: 4x/0,28 (d)
- Filter cubes: 525/50; 609/50 680/42
- Illumination: 488, 561, 647 nm
- Detection: sCMOS: ORCA-Flash 3.0 CAMERALINK; Rolling top
- Resolution: Up to 0.6 um in plane (10X magnification)
- Stage: Motorized (xyz axes + rotation)
- Software: Labview
- Connection: Optical Fiber 10 Gb
- Max size of sample: 5 X 5 X 5 (h) cm
ALICE’s custom-built mesosSPIM microscope, developed in Fritjof Helmchen’s lab at the University of Zurich (UZH), is optimized for large scale screening of cleared samples at cellular resolution with high speed imaging performances.
- Objectives: Detection: Macro-zoom system (Olympus MVX-10) Zoom factor from 0.63X to 4X;
- Illumination: 405, 488, 561, 647 nm
- Filter cubes: 530/43 – LP515 – LP561 – LP663 – 542/27 – 482/35 – 450/50
- Detection: sCMOS: ORCA-Flash 4,0 CAMERALINK; Rolling top
- Resolution: Max resolution: to 1.6 um in plane
- Stage: Motorized (xyz axes + rotation)
- Software: Custom – (Python)
- Max size of sample 5 X 5 X 10 (h) cm
For more information on the mesoSPIM initiative, see mesoSPIM.org
Clearscope, from MBF is light sheet theta microscope designed to meet the unique challenges inherent to imaging at high spatial resolution and allow an unprecedent travel range. This allows our researchers to work with specimens that are too large for traditional light sheet imaging, such as human, non-human primates, and tissue from expansion microscopy.
- Objectives: 4X/0.28 WD 29 mm NA; 20X/1 NA WD 8 mm
- Illumination : 4 lasers channel 405, 488, 561, 640 nm; LSTM with 2 oblique light-sheets; X-Cite LED for widefield detection
- Filter cubes: filter cube with DAPI, TexasRed, FITC, Cy5 and 405/488/561/640 laser QUAD Band; Motorized filter wheel; Reflected light brightfield mirror cube
- Detection: sCMOS: ORCA-Flash
- Resolution: Max resolution: 300 nm in plane
- Stage: Motorized – travel range 150mm x 100mm x 12mm
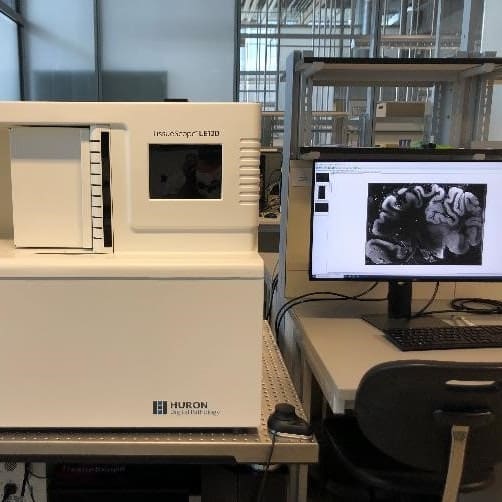
The VS120 is a high-throughput slide scanner that is capable of automatically imaging and stitching up to 100 slides in color bright field mode as well as fluorescence mode. The online stitching allows a fast 2D reconstruction of large sections.
- Objectives: 2X/0.08 WD=6.2, 4X/0.16, 10X/0.4 WD=3.1, 20X/0.75 WD=0.6, 40X/0.9 WD=0.18
- Filter cubes: Ex: 392/23, 474/27, 554/23, 635/15, Em: 433/24, 520/35, 600/37 680/42 nm (motorized turret)
- Illumination: Spectra-X LED
- Detection: B&W CCD, color CCD
- Resolution: Max resolution: 170 nm (40X)
- Stage: Motorized
- Software: VS-ASW
- Slide holders: 100 slides 1” X 3”
TissueScope is a customized automatic slide scanner system for serial 2D fluorescence/BF imaging of large section (scan up to 180×130 mm). Suitable for human brain 2D imaging.
- Objectives: 20X/0.75 WD mm
- Filter cubes: BFP, GFP, Cy3, Cy5
- Illumination: Excelitas_X-Cite_NOVEM – Preview in darkfield
- Detection: Color CMOS 6um pixel 4096 x 3072, Teledyne Photometrics – Kynetics (29.4 mm Field Of View)
- Resolution: Max resolution: 0.3 um
- Stage: Automatic scan and focus control
- Software: Automatic or manual setup and scan, z-scan scan – image format JPEG , TIFF
- Application: Serial and automatized fluorescence/BF microscopy (i.e mosaics of several large sections) – Human Brain
- Slide holders: 120 slides 1” X 3”, 10 custom slide holders
Inverted Widefield microscope DIC, equipped for Brightfield, phase contrast, fluorescence imaging. Excitation up to NIR wavelengths.
- Microscope: Nikon Eclipse Ti2-U partially motorized
- Objectives: Plan Fluor 4x/0.13; Plan Fluor 10x/ 0.3; Plan Fluor 20x/0.5; Plan Fluor 40x /0.75; manual nosepiece, manual condenser
- Filter cubes: BFP, GFP, Cy3, Cy5, Cy7 (motorized turret)
- Illumination: SOLA III V-nIR Light Engine
- Detection: DS-Qi2 Mono Digital Camera
- Stage: Motorized
- Software: NIS Elements BR
Confocal system equipped with 6 laser lines, GaAsP-PMT detectors, Airy Scan detector for multi-channel detection at a resolution higher than traditional confocal. Suitable to any conventional confocal application, including sprectral un-mixing, ratiometric analysis and photobleaching studies. The systems also includes a module for imaging cleared tissue sections or organoids with a clarity optimized objective.
- Objectives: 10x/0,45 (d) WD= 2.0; 20x/0.8 (d) WD= 0.55; 63x/1.4 (oil); 20x/1 (CLARITY), Corr WD= 5.6
- Filter cubes: BFP, GFP, TexasRed
- Illumination: Laser diode 405 nm, Laser Argon Multiline 458, 488, 514 nm; Laser HeNe 543; Laser HeNe 633 nm; Halogen Lamp
- Detection: 1 T-PMT (Transm@ light); 3 ch spectral detection with 1 GaAsP-PMT and 2 PMTs. 1 Airyscan
- Resolution: Max resolution: 140 nm laterally and 400 nm axially
- Stage: Motorized
- Software: Zen 2.3
Spinning disk confocal microscopy utilizes a microlens array disk and a pinhole disk to allow a high-speed 3D imaging of thick sections. The system is optimized for live 3D imaging, thick section 3D and multi-tile imaging, spatial transcriptomics.
- Microscope: Nikon Eclipse Ti2-E fully motorized
- Objectives: Plan Apo Lambda D 10x; Plan Apo Lambda D 20x/0.8; Plan Apo Lambda D 40x /0.95; Plan Apo Lambda D 60x/1.42 Oil; Plan Apo Lambda S 20x/0.95 WI
- Filter cubes: BFP, GFP, Cy3, Cy5, Cy7; (motorized)
- Illumination: Diode laser 405/488/515/561/638/785 120/200/150/200mW with corresponding dichroic (405/488/561/640; 445/515/640; 785); pE-300 LED light source
- Detection: 2X Hamamatsu Orca Fusion BT CoaXPress (22mm FOV, 95% QE, 6.5um pixel size, fast readout, low noise)
- Spinning disk: Spinning Yokogawa CSU W1 – 50um pinhole single – 4000 RPM max speed
- Stage: Motorized
- Software: NIS Elements Advance Research, including JOBS
Contact us for collaborations!
Our technological innovations are driven by strong collaborations with both academic and industry partners. We actively seek new collaborations to push the boundaries of neuroscience, advance our techniques, hardware and software. Reach out to our neuroimaging and neurobiology team for more information.
Partners
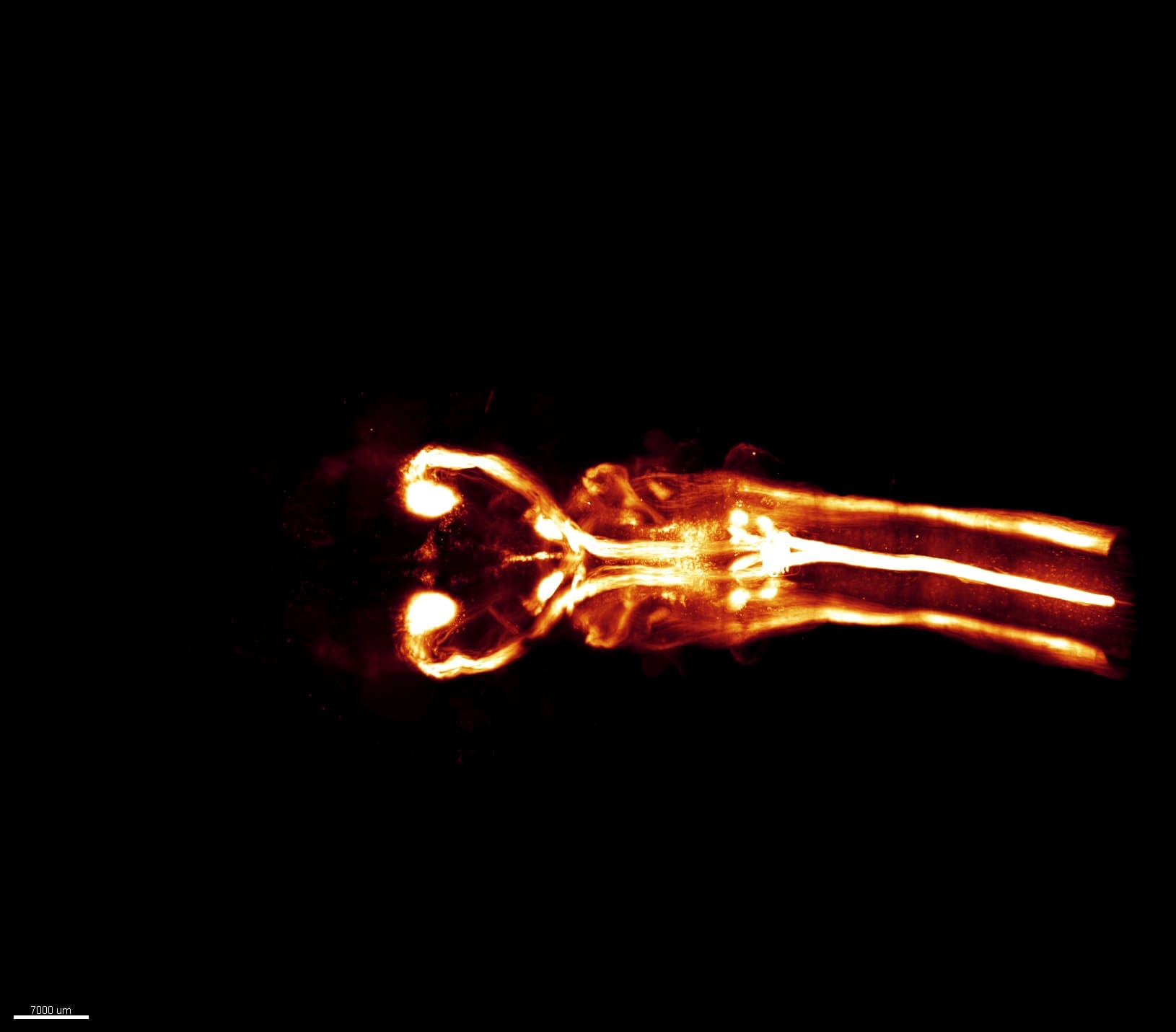
A section of human cerebellum from the Lamylab at the University of Geneva imaged with the Wyss Center’s mesoSPIM lightsheet microscope. The image shows blood vessels labelled with the marker lectin. Blood supplies energy to the brain and is known to be disregulated in some diseases. Imaging brain vasculature with advanced microscopes helps drive research into neurodegenerative diseases and stroke. Credit: Tomàs Jordà – Lamylab, University of Geneva – Human Brain Mapping project

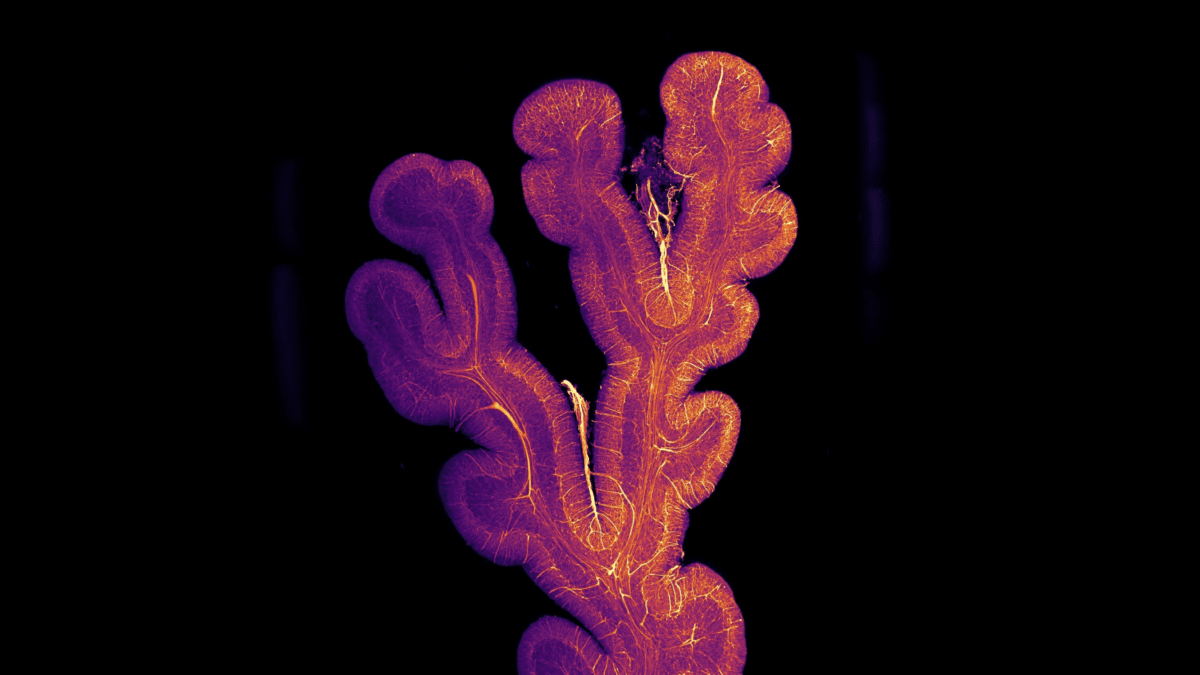
3D image of intestinal villi. Villi are controlled by the enteric nervous system (ENS) our gut’s own little brain. The team is using the three-dimensional microscopic imaging of the intestinal wall to better understand the involvement of enteric neurons in gut function and their connection with the brain. Image acquired with Clarity Optimised Lightsheet Microscope. Gut section cleared with iDisco+ protocol. Data are processed with a depth coding algorithm – the color represents the depth of the 3D image.
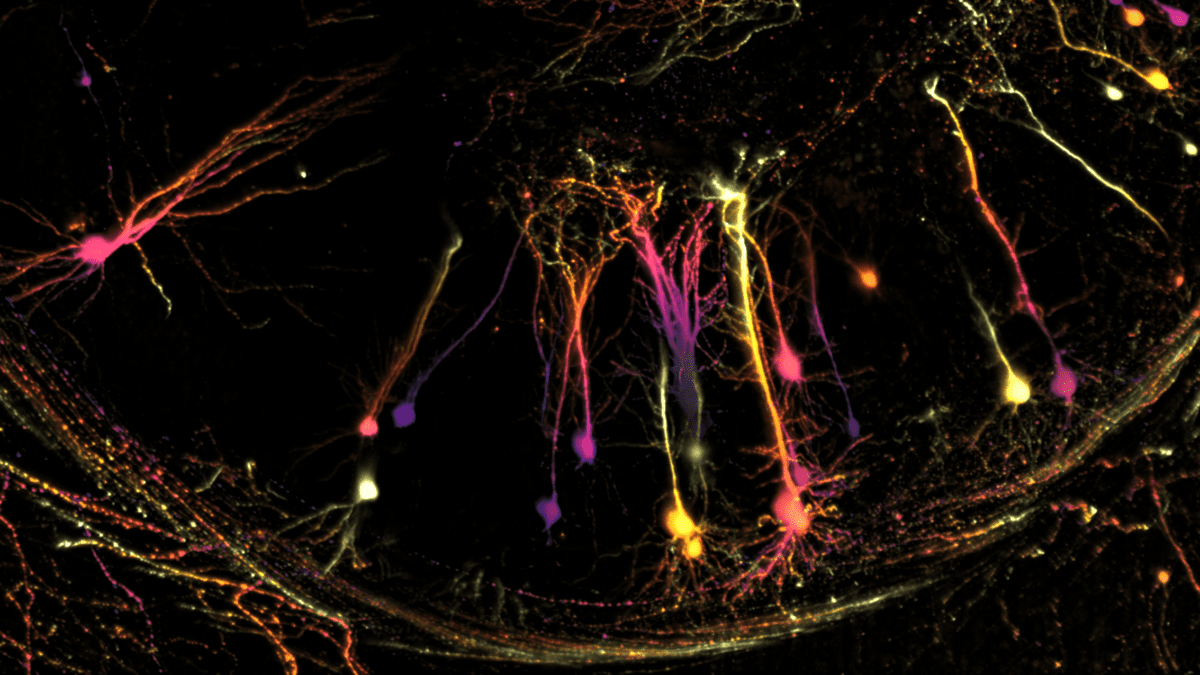
This image of virally labelled neurons in the brain helps reveal the cellular connectivity between the entorhinal cortex and hippocampus. These two regions are involved in memory processing and play a crucial role in the study of neurodegenerative disorders. Image courtesy of Noémie Mazaré, Giovanni Carriero, Roberta de Ceglia, Laura Solanelles, Bilian Xiong, Prof. Ludovic Telley and Prof. Andrea Volterra at the University of Lausanne.
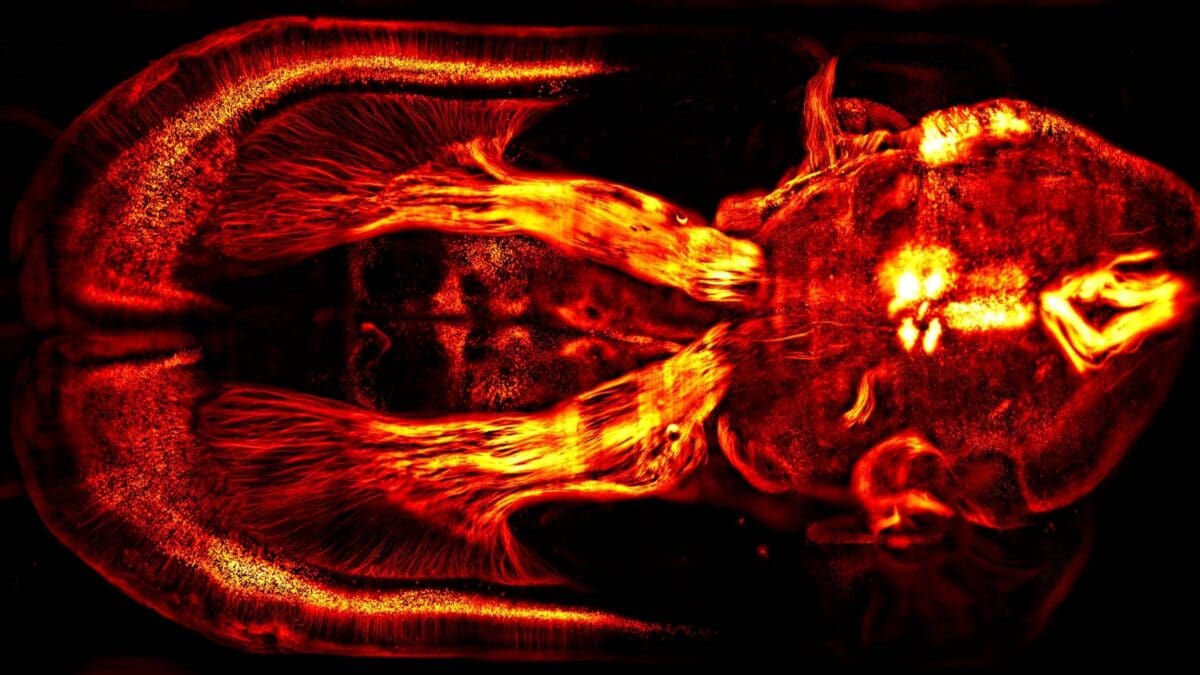
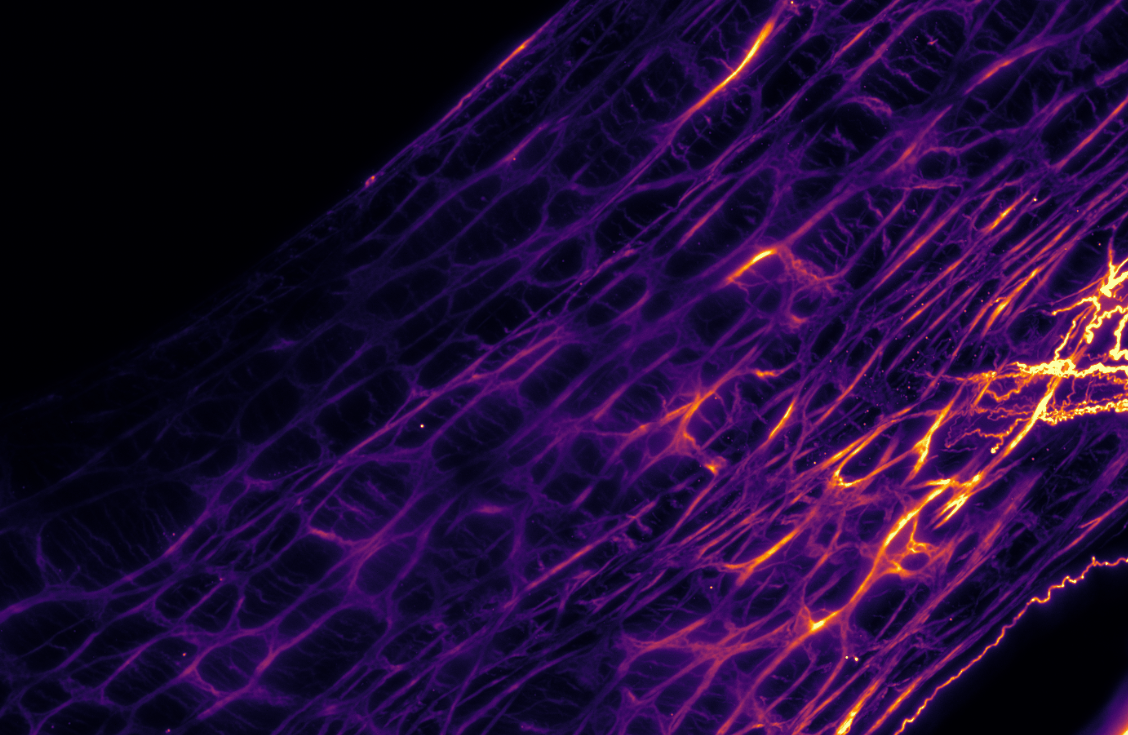
Brain wide labelling of glutamatergic projection neurons – Courtine Lab, EPFL
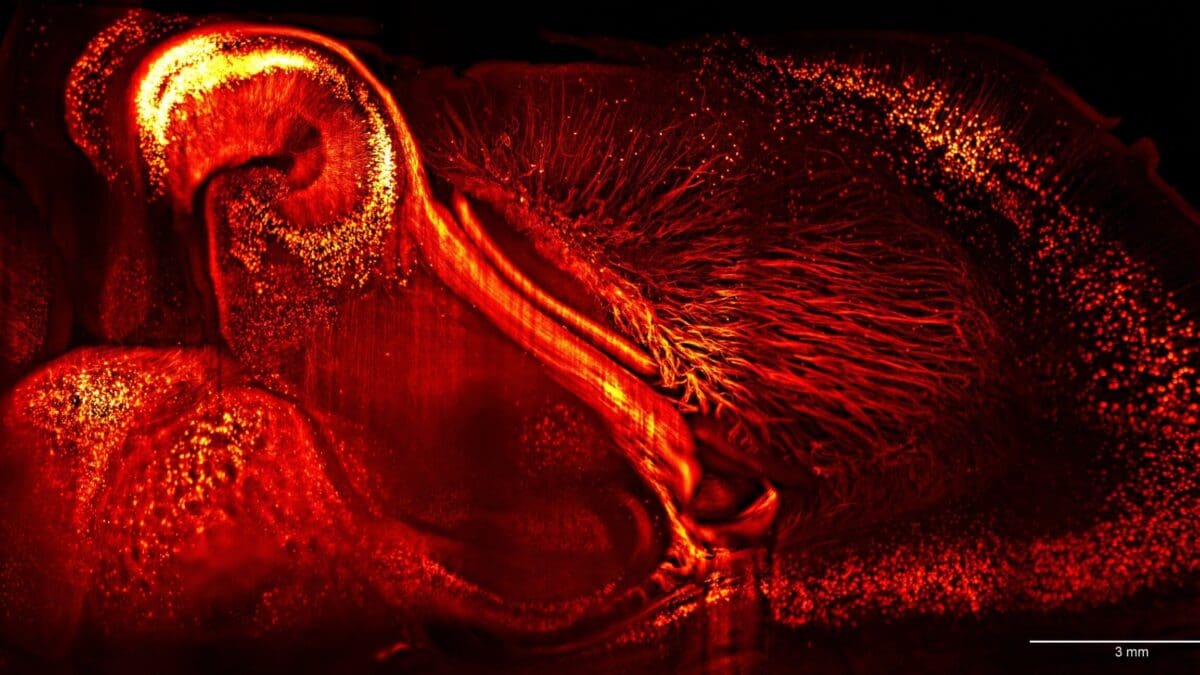
Thy1 GFP, mouse – Holtmaat Lab, University of Geneva

Fluorescent interneurons in a whole brain – Fhyn Lab, University of Oslo