Improving brain tumor treatment
A new view of glioblastoma could help predict patient outcomes

Goal | Understanding the brain tumor microenvironment
Status | Completed
Timeframe | –
Area of Research | Neuro visualization technology
Partners | Wellcome Leap, UNIGE, HUG, Paris Brain Institute
Lead | Stéphane Pagès, Valérie Dutoit
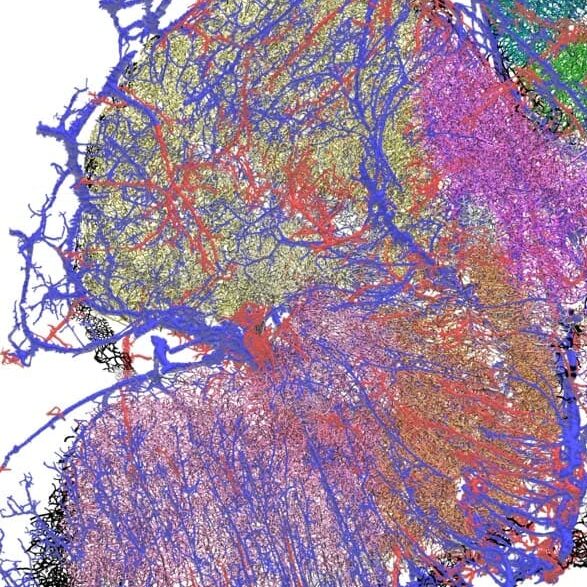
Glioblastoma (GBM) is the most common and aggressive brain tumor, affecting three to five individuals in every 100,000 people. The average age at diagnosis is 60 years and available treatments, involving surgery followed by radio- and chemotherapy, are associated with a median survival of only 15 months. In addition, GBM generally develops resistance to therapy, and only a few patients experience prolonged survival.
The goal of this project is to understand how the tumor microenvironment (TME), particularly immune system components, differ between tumors sampled at diagnosis and at recurrence, and how this correlates with patient response to standard and experimental treatments. In addition, patients with extraordinarily long survival will be analyzed and compared with patients experiencing a shorter course of disease.
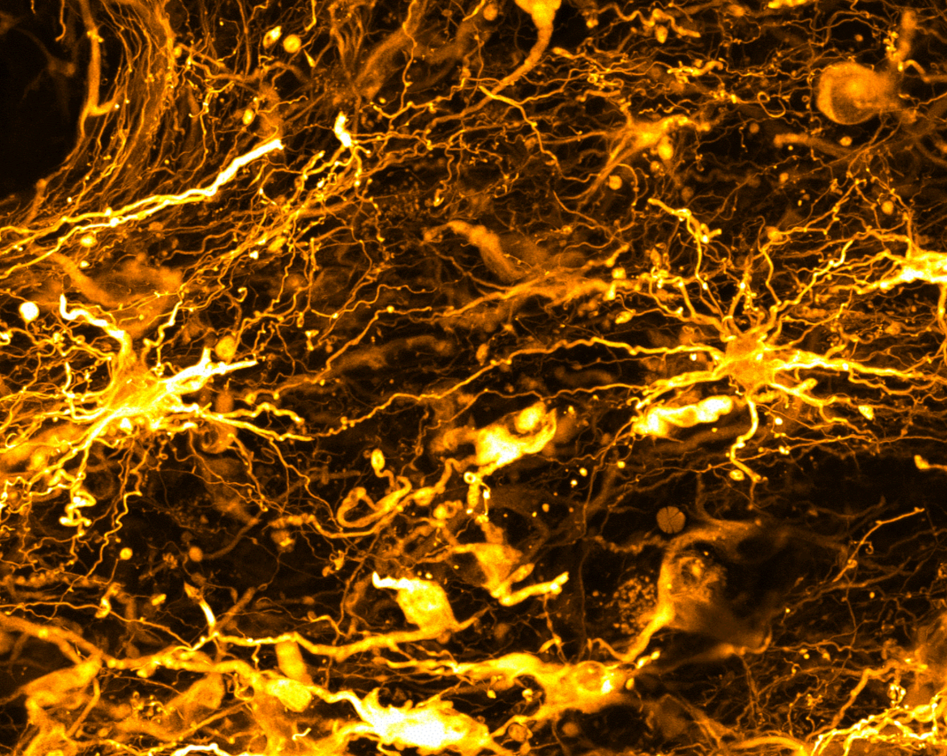
The challenge
Brain tumors are not usually analyzed as a whole. As a result, the complexity of the tumor and its microenvironment is not captured in its entirety.
Investigations are typically performed on very thin GBM slices, only about 10 microns thick, and fail to capture the 3D complexity of the TME. In addition, standard analyses rarely analyze how the TME changes over time upon treatment. GBM comprise many different types of cells, each with their own genetic makeup. This tumor cell diversity enables cells to adapt to their environment. A contributing factor to GBM resistance to current treatments is the immunosuppressive TME composed of immune cells, blood vessels, molecules released by tumor cells as well as hypoxic areas. Altogether, this milieu is immunosuppressive, allowing the tumor to escape the immune response and progress.
A new approach
The team is using high-resolution 3D microscopy to map the spatial distribution of immune cells, tumor cells, genetic markers, blood vessels and hypoxic zones in entire human GBM.
By accurately mapping the TME in tumors from the same patient obtained before and after treatment, the goal is to build a model to help predict response to therapies. In addition, by comparing patients with short versus long survival, it will help identify parameters of the TME associated with longer survival, opening avenues for the development of novel therapies. The experimental pipeline, a series of technologies, not yet applied to human brain tumors, will be used to map the spatial distribution of molecular markers, cells and blood vessels
Tissue Clearing
To enable 3D microscopy of large tumor samples that are millimeters or even centimeters thick, the tissue must be made completely transparent using a process known as clearing.
Clearing removes opaque lipids, including fats, and replaces or modifies them to make the sample transparent.
ELAST
The team is exploring a new method known as entangled link-augmented stretchable tissue-hydrogel (ELAST). This involves embedding the sample in an elastic polymer gel that allows it to deform and reshape. Stretching the sample over hundreds of cycles while it bathes in a solution of antibodies makes labeling of thick samples more efficient.
Labeling
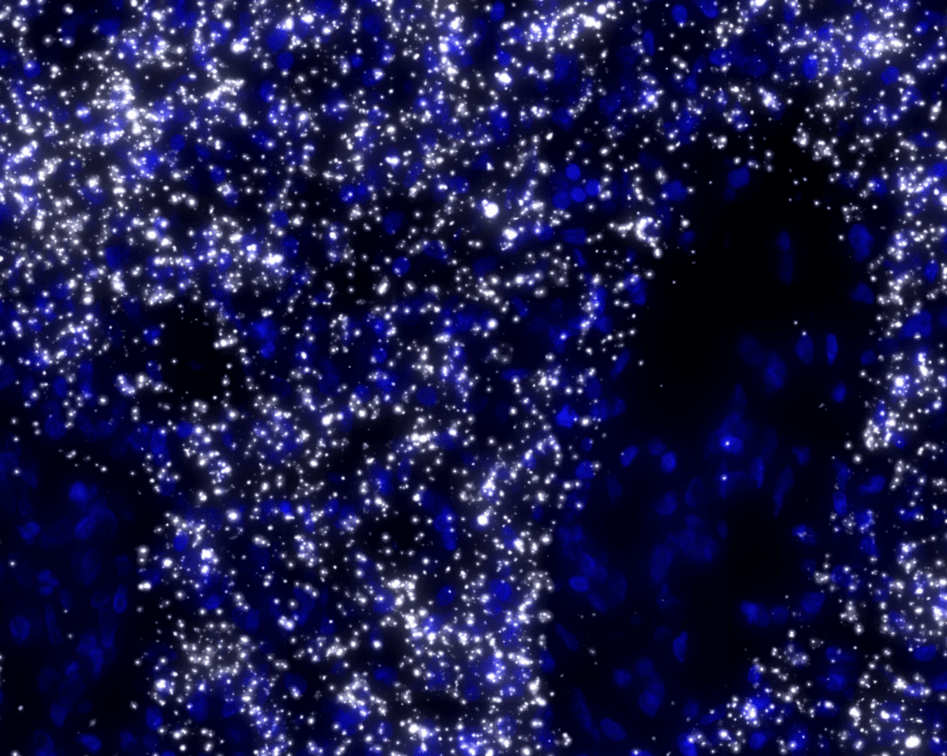
To differentiate between different cell types, blood vessels and other elements of the TME under the microscope, the former are tagged with different fluorescent molecules.
The team uses immunolabeling – a technique in which modified antibodies, that glow under certain wavelengths of light, target proteins of interest. For example, targeting proteins that occur in immune cells allows visualization of these cells throughout the entire sample.
Labeling specific proteins enables the researchers to visualize and understand the interplay between the different components of the tumor and TME.
Spatial transcriptomics
Spatial transcriptomics determines the location of expressed genes within an intact piece of tissue.
The team is using an approach which binds and identifies RNAs from specific genes thought to be involved in tumor development or progression.
This detailed genetic information could be used to target personalized therapies based on the genetics of individual tumors.
Data on the spatial organization of molecular markers, cell populations and vasculature in a tumor will be integrated with clinical data. Together, the information will be used to train models and predict tissue states over time.
Ultimately, the goal is to be able to understand what parameters are modified over time upon treatment and are possibly associated with better outcomes. This could help optimize treatment for patients or help to develop novel treatment modalities.
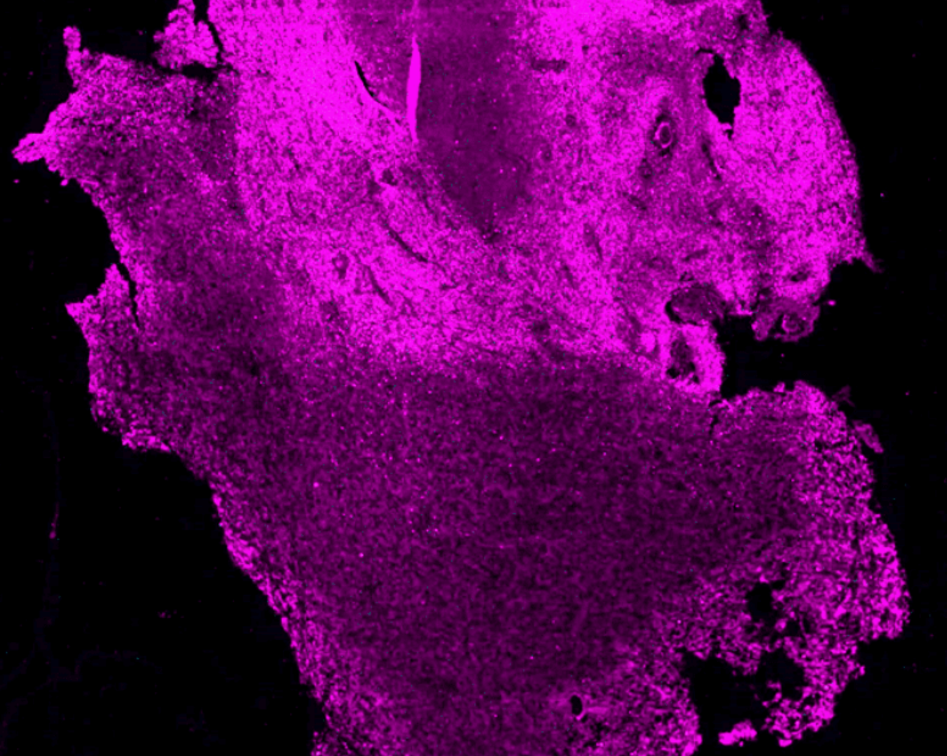
Experimental therapies
The local immunosuppressive effect of the GBM TME is thought to play a role in tumor progression. A new generation of immunotherapies attempts to reverse this immunosuppression by boosting the immune response. However, these strategies do not yet benefit the majority of patients.
A phase I/II clinical trial testing a GBM-derived peptide vaccine combined with an immune checkpoint inhibitor is enrolling patients with recurrent GBM at the University of Geneva and Geneva University Hospital. This immunotherapy trial aims at stimulating the patient’s immune system, and in particular T cells, towards tumor cells.
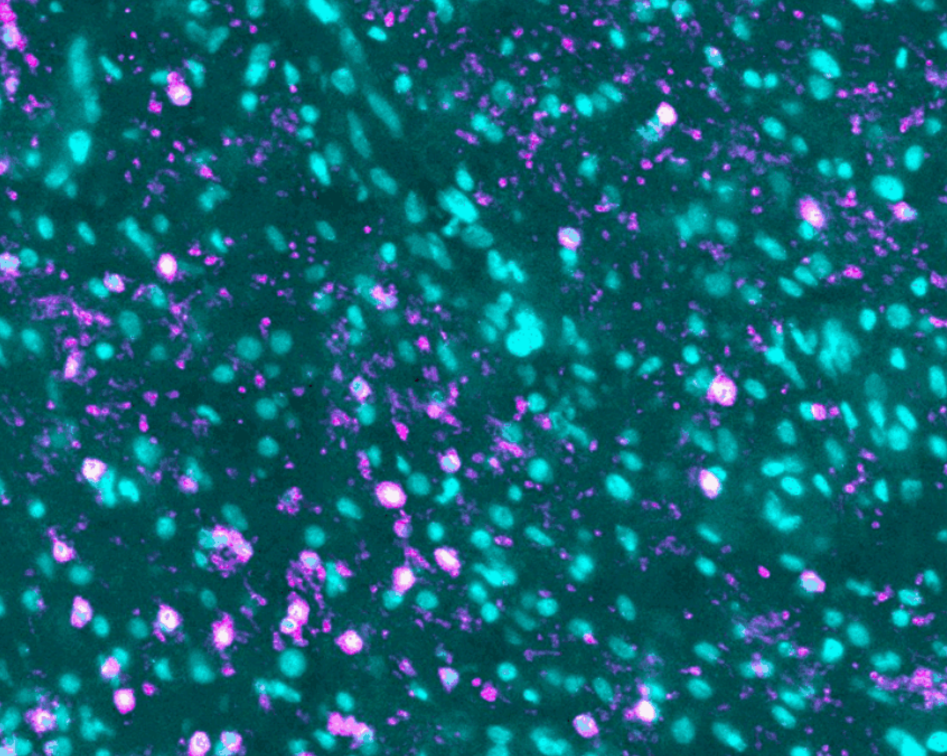
The team will map tumors from two patient cohorts in the trial, one being treated with the peptide vaccine alone and one with the vaccine combined with the immune checkpoint inhibitor pembrolizumab, an antibody used to potentiate vaccine-elicited T cell responses. This will help understand the effects of the peptide vaccine on the GBM TME and potentially identify parameters associated with patient outcome.